
M.S. - 1983 - Indiana University School of Medicine
Ph.D. - 1988 - University of Utah
Postdoc - 1988-89 - University of California, San Francisco
Laboratory is full until January 2025
Dr. Davisson's research has historically covered a broad range of experimental approaches, from mechanism-based drug design to analytical instrument testing and development. He has formal graduate education and training in organic chemistry and biochemistry and has been called a pioneer in the now-maturing arena of chemical biology. His interdisciplinary efforts continue to guide and motivate the current research program. His continued efforts to develop multidisciplinary education and training environments are represented by his long-standing commitment to the academic unit's mission.
The general interests are at the intersection of chemistry and disease biology to enhance the drug discovery and development process. For this reason, the laboratory has engaged in the early stages of development for different platform technologies over many years.
Currently, the research group uses hypothesis-driven and technology-focused discovery approaches to address therapeutic strategies for unmet needs in treating several cancer diseases, emerging viral infections, and neurodegenerative diseases.
Our character is to engage in collaborative efforts to enhance the overall approaches to addressing these objectives and promote the translation to the clinic.
While the first 30 years of research focused primarily on metabolic enzymatic targets, all of the molecular systems under the current investigation are considered non-classical or “undruggable” targets. Our efforts aim to discover and develop small molecule probes of these target systems to address their specific roles in disease contexts and serve as leads for drug discovery. A significant effort is devoted to exploring new approaches to designing and discovering useful chemotypes and drug leads for each target system. We develop probes to test hypotheses regarding protein network interactions and define new target binding sites. Currently, biomolecular screening methods are integrating with computational approaches and novel synthetic chemical libraries to enhance the success of the discovery process.
The general themes of the current research are:
- Chemical probes & medicinal chemistry for antivirals, anticancer, neurodegeneration
- Drugging non-conventional protein targets
- New modalities for drug development beyond the rule of 5 molecules
Our active core research programs can be stated in 3 broad areas.
1) To discover and develop selective antagonists/agonists of protein assemblies relevant to therapeutic cancer interventions.
The current target systems under investigation involve various cellular roles, including
- DNA replication and repair
- cellular vesicle transport and pH control
- viral-mediated oncogenesis
The specific project targets currently under investigation include:
- cell proliferating nuclear antigen (PCNA)
- human papillomavirus virus E6 protein (HPV-E6)
- nuclear localized EGFR and DNA-PK
2) To harness chemical scaffolds and mechanisms of action of natural product drugs.
A long-standing interest is in understanding molecular mechanisms of drug actions. Part of the inspiration comes from the rich biological activities of natural products and their synthetic analogs, which have the potential to be developed into new drug therapies.
Natural products as molecular tools continue to provide rich sources for drug target discovery and/or serve as starting point scaffolds for designing new therapeutics. We continue to pursue biochemical/proteomic and biophysical/structural biology approaches to understand and exploit the cellular pharmacology of natural products in future drug design.
The current projects focus on:
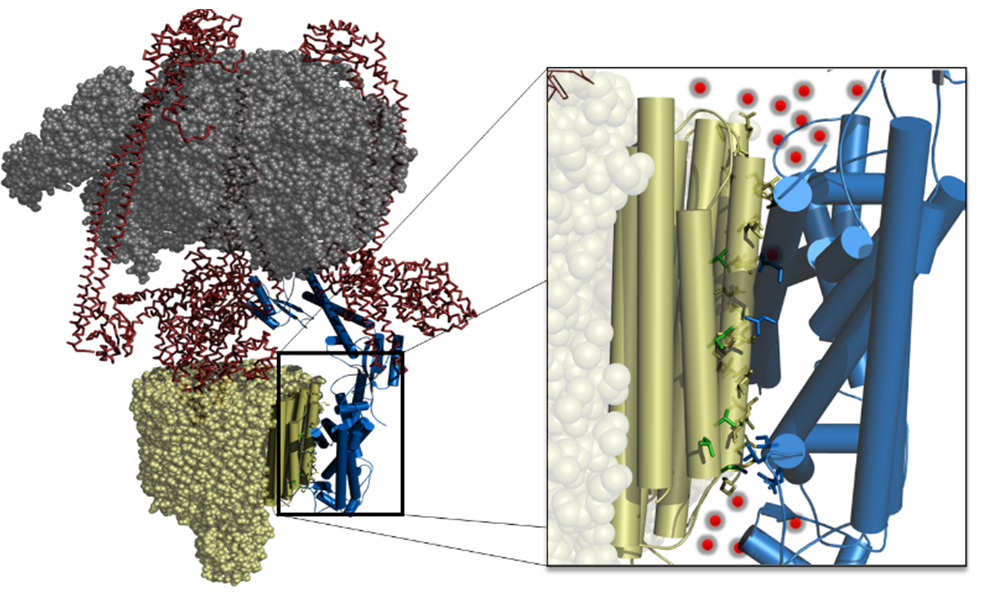
- Natural product scaffolds that modulate the function of vacuolar-ATPase (v-ATPase) for drug development as antivirals and anti-metastatic activities.
- The use of ligand-based and structure-based drug design approaches to discover new and improved selective modulators of V-ATPase isoforms
3) To develop or utilize novel high-content, quantitative, phenotypic-based screens for molecular discovery and evaluation.
These collaborative efforts involve platform development bridging with chemical biology using bioengineering and data sciences approaches.
The variation of biological response to chemical effects as a function of genetic content in biological systems is a problem for integrating genomics and proteomics with high-content phenotypic assay systems. Our collaborative efforts aim to use innovative biosystem screens to evaluate new agents' efficacy, toxicity, or pharmacokinetic properties. Insights from these efforts offer an understanding of how best to target susceptibilities and stage drug therapies from discovery through development.
The current projects involve:
- The use of zebrafish models to guide the development of novel toxicant countermeasures with a current focus on treatments for cyanide exposure
- Molecular interventions for mitochondrial dysfunctions to counteract neurodegenerative diseases.
Naoaki Fujii (Principal Research Scholar)
Payton Martin (Research Technician)
Ethan K. Miyake (Lab Research Technician)
Laura Mae Sanford (Graduate Student)
Vallabh Suresh (Post-Doctoral Research Associate)
Hui Tao (Graduate Student)
Charlie C. Zhang (Senior Research Associate)
Drug discovery and design technologies
- Chemical probes & medicinal chemistry for antivirals, anticancer, neurodegeneration
- Drugging non-conventional protein targets
- New modalities for drug development beyond the rule-of-5 molecules
Diseases:
- Cancers: breast cancers, pancreatic cancer, HPV-associated cancers
- Emerging infectious agents: coronavirus, ebola virus, dengue virus.
- Degenerative diseases: ALS, Parkinsons, Alzheimer's, chronic kidney disease
- Cyanide and related toxicant exposures
Technology commercialization
- new chemical sensing agents for molecular detection in clinical diagnostics
- activity biosensing technologies for biomarker diagnostics
- new molecular entities as antiviral therapeutics
- molecularly targeted adjuvant agents for cancer therapies
Promoting entrepreneurship in pharmaceuticals and medical diagnostics
Undergraduates in academic research
- ACS Medicinal Chemistry Division Long Range Planning 2017-2019
- NIH Review panels
- Founder and Chief Scientific Officer of Amplified Sciences
MCMP 570 Basic Principles of Chemical Action on Biological Systems
PHRM 460 Drug Discovery and Development I
1978 American Institute of Chemists Undergraduate Award
1986 University of Utah Graduate Student Research Prize
1989 Damon Runyon-Walter Winchell Fellowship
1999 Purdue University Scholar
2004 Walther Cancer Institute Research Prize
2007 Purdue Cancer Center Lions Club Award
2012 Fellow of American Association for Advancement of Science
2014 Chaney Research Scholar Award
2018 & 2022 Teacher of the Year Award for BS Pharmaceutical Sciences
B.S. Pharmaceutical Sciences Oversight Committee
Work Force of the Future in Pharmaceutical Manufacturing
Jeffrey D. Altenburg, Andrew A. Bieberich, Colin Terry, Kevin A. Harvey, Justin F. VanHorn, Zhidong Xu, V Jo Davisson and Rafat A. Siddiqui (2011) “A synergistic antiproliferation effect of curcumin and docosahexaenoic acid in SK-BR-3 breast cancer cells: unique signaling not explained by the effects of either compound alone” BMC Cancer 11, 149.
Qingshou Chen, Dirk Schweitzer, John Kane, V. Jo Davisson, and Paul Helquist (2011) “Total synthesis of iejimalide B” J. Org. Chem., 76, 5157–5169.
Mout, R., Xu, Z-D., Wolf, A., Davisson, V.J., Jarori, G.K. (2012) “Anti-malarial activity of geldanamycin derivatives in mice infected with Plasmodium yoelii” Malaria Journal 11: 54
Robinson, J.P., Rajwa, B., Patsekin, V., Davisson, V. J. (2012) “Computational analysis of high-throughput flow cytometry data” Exp Opin Drug Discovery 7 (8) 679-693.
Tsiper, M. V., Sturgis, J., Avramova, L. V., Parakh, S., Fatig, R., Juan-García, A., Li, N., Rajwa, B., Narayanan, P., Qualls, Jr., C. W., Robinson, J. P., Davisson, V. J. (2012) “Differential Mitochondrial Toxicity Screening and Multi-parametric Data Analysis” PlosOne 7 (10) e45226
D'Ordine RL, Linger RS, Thai CJ, Davisson V. J. “Catalytic zinc site and mechanism of the metalloenzyme PR-AMP cyclohydrolase.” (2012) Biochemistry 51(29):5791-803.
Han, B., Wright, R., Kirchhoff, A. M., Chester, J. A., Cooper, B. R., Davisson, V. J., Barker, E. (2013) "Quantitative LC-MS/MS analysis of arachidonoyl amino acids in mouse brain with treatment of FAAH inhibitor" Anal. Biochem. 432 (2):74-81.
Robinson, J. P., Holdman, C., Ragheb, K., Sturgis, J., Fatig, R., Avaramova, L. V., Rajwa, B., Davisson, V. J., Lewis, N., Narayanan, P., Li, N., Qualls Jr., C. W. (2013) “High-Throughput Secondary Screening at the Single-Cell Level” J. Lab Automation (1):85-98.
Oliver, J. S., Linger, R. S., Chittur, S. V., Davisson, V. J. “Substrate Activation and Conformational Dynamics of Guanosine 5′-Monophosphate Synthetase” (2013) Biochemistry, 52, 5225−5235
Siddiqui R. A., Harvey K. A., Xu Z., Natarajan S. K., Davisson V. J. (2014) “Characterization of lovastatin-docosahexaenoate anticancer properties against breast cancer cells” Bioorg Med Chem. 22(6):1899-908. doi: 10.1016/j.bmc.2014.01.051 PMID: 24556504
Oliver J. C., Gudihal R., Burgner J. W., Pedley A. M., Zwierko A. T., Davisson V. J., Linger R. S. (2014) “Conformational changes involving ammonia tunnel formation and allosteric control in GMP synthetase” Arch Biochem Biophys. 545:22-32. doi: 10.1016/j.abb.2014.01.004. PMID: 24434004
Pedley, A. M., Lill, M. A., Davisson, V. J. (2014) “Flexibility of PCNA-Protein Interface Accommodates Differential Binding Partners” PlosOne e102481. doi: 10.1371/journal.pone.0102481 PMID:25036435.
Bartolowits M, Davisson V. J. (2015) “Considerations of Protein Subpockets in Fragment-Based Drug Design” Chem Biol Drug Des. Jan;87(1):5-20. doi: 10.1111/cbdd.12631. Epub 2015 Aug 31. PMID: 26307335
Rietz A., Petrov D. P., Bartolowits M., DeSmet M., Davisson V. J., Androphy E. J. (2016) Molecular Probing of the HPV-16 E6 Protein Alpha Helix Binding Groove with Small Molecule Inhibitors. PLoS One :e0149845. doi: 10.1371/journal.pone.0149845. PMID: 26915086
Thomas F.M., Goode K.M., Rajwa B., Bieberich A.A., Avramova L.V., Hazbun T.R., Davisson V.J. (2017) “A Chemogenomic Screening Platform Used to Identify Chemotypes Perturbing HSP90 Pathway” SLAS Discov. Jul;22(6):706-719. doi: 10.1177/2472555216687525. Epub 2017 Jan 31. PMID:28346089
Goode K.M., Petrov D.P., Vickman R.E., Crist S.A., Pascuzzi P.E., Ratliff T.L., Davisson V.J., Hazbun T.R. (2017) “Targeting the Hsp90 C-terminal domain to induce allosteric inhibition and selective client downregulation” Biochim Biophys Acta. Aug;1861(8):1992-2006. doi: 10.1016/j.bbagen.2017.05.006. Epub 2017 May 8. PMID:2849520
Bartolowits M.D., Brown W., Ali R., Pedley A.M., Chen Q., Harvey K.E., Wendt M.K., Davisson V.J. (2017) “Selective Inhibition of STAT3 Phosphorylation Using a Nuclear-targeted Kinase Inhibitor” ACS Chem Biol. 2017 Aug 8. doi: 10.1021/acschembio.7b00341. [Epub ahead of print] PMID:28787571
Sips PY, Shi X, Musso G, Nath AK, Zhao Y, Nielson J, Morningstar J, Kelly AE, Mikell B, Buys E, Bebarta V, Rutter J, Davisson VJ, Mahon S, Brenner M, Boss GR, Peterson RT, Gerszten RE, MacRae CA. (2018) “Identification of specific metabolic pathways as druggable targets regulating the sensitivity to cyanide poisoning.” Plos One 13 (6):e0193889. doi: 10.1371/journal.pone.0193889. PMID:29879736
Ali R, Brown W, Purdy SC, Davisson VJ, Wendt MK. (2018) “Biased Signaling Downstream of Epidermal Growth Factor Receptor Regulates Proliferative versus Apoptotic Response to Ligand” Cell Death Dis. 9 (10):976. doi: 10.1038/s41419-018-1034-7. PMID:30250119
Lindstrom A, Anantpadma M, Baker L, Raghavendra NM, Davey R, Davisson VJ. (2018) “Phenotypic Prioritization of Diphyllin Derivatives That Block Filoviral Cell Entry by Vacuolar (H+)-ATPase Inhibition.” ChemMedChem. 13 (24):2664-2676. doi: 10.1002/cmdc.201800587. PMID:30335906
Bartolowits, M. D., Xin, M., Petrov, D., Tague, T., Davisson, V. J. (2019) “Multimeric Rhodamine Dye-Induced Aggregation of Silver Nanoparticles for Surface-Enhanced Raman Scattering” ACS Omega 4 (1) 140-145. doi:10.1021/acsomega.8b02970. PMID: 30729221
Morningstar J, Lee J, Hendry-Hofer T, Witeof A, Lyle LT, Knipp G, MacRae CA, Boss GR, Peterson RT, Davisson VJ, Gerszten RE, Bebarta VS, Mahon S, Brenner M, Nath AK. (2019) “Intramuscular administration of hexachloroplatinate reverses cyanide-induced metabolic derangements and counteracts severe cyanide poisoning.” FASEB Bioadv. 2019 Feb;1(2):81-92. doi: 10.1096/fba.1024. Epub 2018 Oct 8. PMID: 31355359
Bartolowits M. D., Gast J. M., Hasler A. J., Cirrincione A. M., O'Connor R. J., Mahmoud A. H., Lill M. A., Davisson V. J. (2019) “Discovery of Inhibitors for Proliferating Cell Nuclear Antigen Using a Computational-Based Linked-Multiple-Fragment Screen.” ACS Omega Sep 6;4(12):15181-15196. doi: 10.1021/acsomega.9b02079. eCollection 2019 Sep 17 PMID: 31552364
Nielson JR, Nath AK, Doane KP, Shi X, Lee J, Tippetts EG, Saha K, Morningstar J, Hicks KG, Chan A, Zhao Y, Kelly A, Hendry-Hofer TB, Witeof A, Sips PY, Mahon S, Bebarta VS, Davisson VJ, Boss GR, Rutter J, MacRae CA, Brenner M, Gerszten RE, Peterson RT. (2022) “Glyoxylate protects against cyanide toxicity through metabolic modulation.” Sci Rep. (2022) 12 (1):4982. doi: 10.1038/s41598-022-08803-y PMID: 35322094.
Plescia CB, Lindstrom AR, Quintero MV, Keiser P, Anantpadma M, Davey R, Stahelin RV, Davisson VJ. (2022) “Evaluation of Phenol-Substituted Diphyllin Derivatives as Selective Antagonists for Ebola Virus Entry.” ACS Infect Dis. 8 (5):942-957. doi: 10.1021/acsinfecdis.1c00474. PMC9112336
Behymer MM, Mo H, Fujii N, Suresh V, Chan A, Lee J, Nath AK, Saha K, Mahon SB, Brenner M, MacRae CA, Peterson R, Boss GR, Knipp GT, Davisson VJ. (2022) “Identification of Platinum(II) Sulfide Complexes Suitable as Intramuscular Cyanide Countermeasures.” Chem Res Toxicol. 35 (11):1983-1996. doi: 10.1021/acs.chemrestox.2c00157. PMID: 36201358
Suresh V, Byers K, Rajesh UC, Caiazza F, Zhu G, Craik CS, Kirkwood K, Davisson VJ, Sheik DA. (2022) “Translation of a Protease Turnover Assay for Clinical Discrimination of Mucinous Pancreatic Cysts” Diagnostics (Basel) 12 (6):134 doi: 10.3390/diagnostics12061343 PMID: 35741154
Bebarta VS, Shi X, Zheng S, Hendry-Hofer TB, Severance CC, Behymer MM, Boss GR, Mahon S, Brenner M, Knipp GT, Davisson VJ, Peterson RT, MacRae CA, Rutter J, Gerszten RE, Nath AK. (2023) “Intramuscular Administration of Glyoxylate Rescues Swine from Lethal Cyanide Poisoning and Ameliorates the Biochemical Sequalae of Cyanide Intoxication.” Toxicol Sci. Nov 3;. doi: 10.1093/toxsci/kfac116. PMID: 36326479
Waiker DK, Verma A, Akhilesh, A GT, Singh N, Roy A, Dilnashin H, Tiwari V, Trigun SK, Singh SP, Krishnamurthy S, Lama P, Davisson VJ, Shrivastava SK. (2023) “Design, Synthesis, and Biological Evaluation of Piperazine and N-Benzylpiperidine Hybrids of 5-Phenyl-1,3,4-oxadiazol-2-thiol as Potential Multitargeted Ligands for Alzheimer's Disease Therapy” ACS Chem Neurosci. 14(11):2217-2242. doi: 10.1021/acschemneuro.3c00245. PMID:37216500
Behymer MM, Mo H, Fujii N, Suresh V, Arzumanian AS, Chan A, Nath AK, McCain R, MacRae CA, Peterson R, Boss GR, Davisson VJ, Knipp GT (2023) “Investigating the Replacement of Carboxylates with Carboxamides to Modulate the Safety and Efficacy of Platinum(II) Thioether Cyanide Scavengers” Toxicol Sci. Nov 11;197(2):197-210. doi: 10.1093/toxsci/kfad119. PMID: 37952247

